Introduction
Plant viruses are one of major threat pathogens and causing serious economic losses to the crops. Although plant viruses have simple genetic information, its interactions with plants is largely still unclear. To control viral diseases, several approaches have been used such as crop rotation, destruction of infected sources, breeding for resistance, and chemical control of relevant insect vectors (Hull, 2002)(1). In nature, plants are constantly exposed to viruses. To survive from viruses, plants have developed sophisticated defense mechanisms during the plant-virus interactions. However, interestingly most plants are resistant to plant viruses, which suggests that susceptibility to viral pathogens is the exception rather than the rule.
One of the well-characterized defense mechanism is R protein-mediated resistance, which implys that R protein interacts with its race specific corresponding avirulence (Avr) genes directly or indirectly to activate defense signaling and suppresses growth of pathogens. (“gene-for-gene” interaction [Flor, 1971])(2). This R-Avr interaction triggers at least one defense signaling cascades and is often associates with HR (Hypersensitive Response), a form of programmed cell death, at the site of pathogen entry (Dangl and Jones, 2001)(3), which trigger high expression of defense genes including pathogenesis-related (PR) proteins (Fig. 1). However, increasing evidence suggesting that the HR is not a essential factor for gene-for-gene mediated disease resistance. For example, a visible HR does not develop either in potato during Rx-mediated resistance against Potato virus X (PVX) (Chandra-Shekara et al., 2004)(4). Thus, while an HR is frequently associated with resistance, it is not necessarily required for resistance.
R-protein mediated defense response also associates with various plant hormones such as salicylic acid (SA), jasmonic acid (JA), ethylene (ET), which in turn signal the activation of defense gene expression. These hormones regulate defense signaling pathways individually, synergistically or antagonistically (Kachroo et al., 2000)(5).
R genes cloned to date have been grouped into six classes regards of their structural characteristics (Martin et al., 2003)(6). Interestingly, most of R genes so far cloned have nucleotide binding site (NBS)-leucine-rich repeats (LRR) type R proteins predominately. Among them, several R genes that confer resistance to plant viruses have been cloned and characterized (Soosaar et al., 2005)(7). Most plant virus resistance genes encoding coiled-coil (CC) or toll-interleukin-1 receptor (TIR) at N-terminus and NBS and LRR domains (Table 1).
Considerable progress had been investigated to understand how plants defend themselves and the mechanisms resistance to viruses (Zaitlin M et al., 2000)(21). However, viral pathogens are still one of major threat and the understanding of viral resistance mechanism is still open question and certainly, more work is needed to elucidate the complex signaling cascade leading to resistance. Turnip crinkle virus (TCV)-Arabidopsis pathosystem is one of the few manipulable plant-virus systems in which resistance is conferred via pathogen- induced defense responses and where HR and resistance can be studied as two distinct phenotypes. The TCV-Arabidopsis pathosystem also provides various other advantages for studying compatible plant-virus interactions. This review covers recent advances on TCV-Arabidopsis pathosystem as a model of virus resistance signaling pathway, as well as the mechanism underlying virus resistance, and its future perspectives.
| R gene | Host* | Pathogen† | Avr factor‡ | Predicted domains§ | Reference |
|---|---|---|---|---|---|
| N | N. tabacum | TMV | Helicase | TIR-NBS-LRR | Whitham et al., 1994(8) |
| HRT | A. thaliana | TCV | CP | CC-NBS-LRR | Cooley et al., 2000(9) |
| RCY1 | A. thaliana | CMV | CP | CC-NBS-LRR | Takahashi et al., 2002(10) |
| RTM1 | A. thaliana | TEV | -¶ | Jacaline-like | Chisholm et al., 2000(11) |
| RTM2 | A. thaliana | TEV | - | Small HSP-like | Whitham et al., 1994(8) |
| Rχ1 | S. tuberosum | PVX | CP | CC-NBS-LRR | Bendahmane et al., 1999(12) |
| Rχ2 | S. tuberosum | PVX | CP | CC-NBS-LRR | Bendahmane et al., 2000(13) |
| Nb | S. tuberosum | PVX | MP | - | Marano et al., 2002(14) |
| Nytbr | S. tuberosum | PVY | - | - | Celebi-Toprak et al., 2002(15) |
| Ry | S. tuberosum | PVY | NIaPro | - | Mestre et al., 2003(16) |
| Sw-5 | L. esculentum | TSWV | - | CC-NBS-LRR | Brommonschenkel et al., 2000(17) |
| Tm-1 | L. esculentum | ToMV | RdPp | - | Meshi et al., 1988(18) |
| Tm-2 | L. esculentum | ToMV | MP | CC-NBS-LRR | Lanfermeijer et al., 2005(19) |
| Tm-22 | L. esculentum | ToMV | MP | CC-NBS-LRR | Lanfermeijer et al., 2005(19) |
| Rsv1 | G. max | SMV | HC-pro and P3 | CC-NBS-LRR | Hayes et al., 2004(20) |
*A. thaliana, Arabidopsis thaliana; N. tabavum, Nicotiana tabacum; S. tuberosum, Solanum tuberosum; L. esculentum, Lycopersicum esculentum; G. max, Glycine max.
†TCV, Turnip crinkle virus; CMV, Cucumber mosaic virus; TMV, Tobacco mosaic virus; PVX, Potato virusX; PVY, Potato virus Y; TSWV, Tomato spooted wilt virus; ToMV, Tomato mosaic virus; SMV, Soybean mosaic virus; TEV, Tobacco etch virus.
‡CP, coat protein; MP, movement protein; RdRp, RNA dependent RNA polymerase; NIaPro, protease domain of unclear inclusion; HC-Pro, helper component protease.
Plant defense signaling to TCV
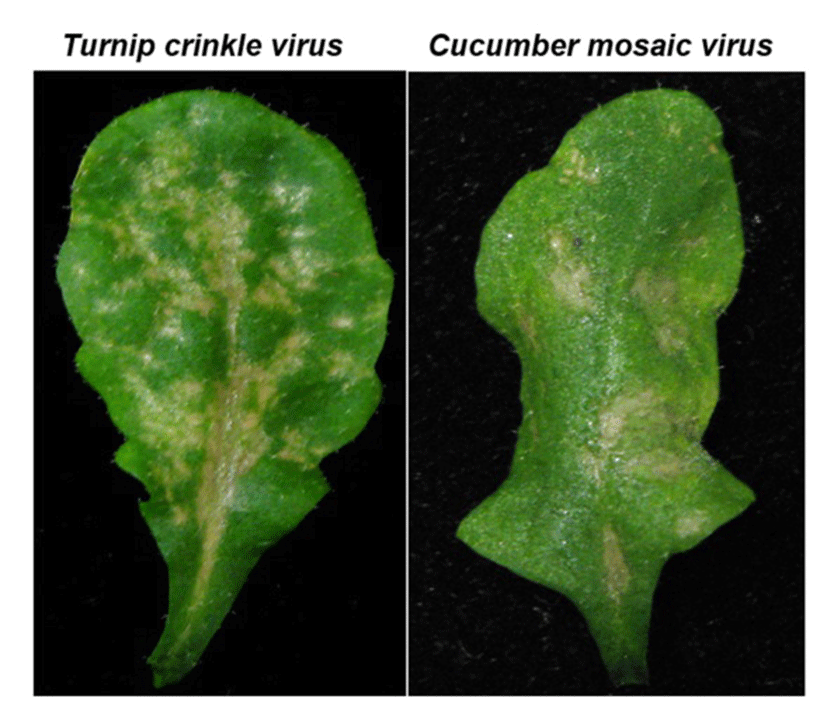
TCV belongs to the icosahedral Carmovirus genus in Tombusviridae family. TCV is a single- stranded positive RNA virus encoding five open reading frames which are required for virus replication (p28 and p88), movement (p8 and p9), encapsidation and movement (p38, coat protein) and its genome size is 4 kb. The TCV has a broad host range that includes several members of the cruciferous family and shows distinquishable systemic symptoms, such as mottling, leaf distortion and crinkling, chlorotic lesions and stunting (Hacker et al., 1992)(22). Most Arabidopsis ecotypes are susceptible to TCV; however, a resistant line, designated Di-17 was isolated from the Dijon (Di) ecotype is resistant to TCV (Dempsey et al., 1993)(23). Following TCV infection, Di-17 plants develop a HR, express several defense genes, including PR (Pathogenesis Related)-1, PR-2, PR-5 and GST1 (Glutathione S-transferase 1) and accumulate SA and phytoalexin, camelexin (Kachroo et al., 2000; Dempsey et al., 1993)(5, 23). By contrast, susceptible plants, lacking HRT, fail to develop an HR after TCV infection, they also exhibit basal level expression of the PR and GST1 genes and accumulate low levels of SA. These susceptible plants allow systemic spread of TCV in uninoculated leaves and finally show crinkled leaves and drooping blots symptoms (Fig. 2).
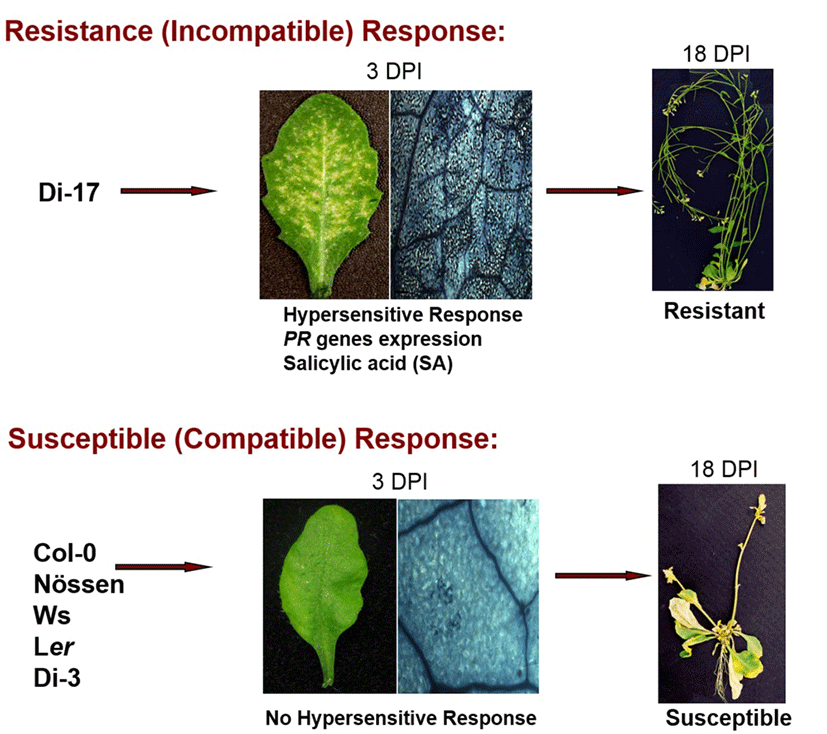
HR and Resistance to TCV is conferred by the dominant resistance protein, designated HRT (hypersensitive response to TCV), which is a CC-NBS-LRR type R protein and show identity to its paralogs RPP8 and RCY1 conferring resistance to Hyaloperonospora arabidopsidis Emco5 and Cucumber mosaic virus in Arabidopsis, respectively. HRT-mediated resistance signaling is activated in the presence of TCV coat protein (CP) as an avirulence determinant (Jeong et al., 2008)(24). However, direct interactions between HRT and CP have not been detected. HRT alone is not sufficient for TCV resistance, because all F1 plants and ~75% of F2 plants derived from a cross between resistant (Di-17) and susceptible (Col-0) plants show symptoms to TCV. Moreover, more than 90% of Col-0 plants expressing HRT gene develop normal HR but are still susceptible, suggesting that other loci also might be regulating resistance to TCV. Following studies revealed that the recessive allele of a second locus, as yet unidentified, designated rrt (regulates resistance to TCV), is also required for TCV resistance (Chandra-Shekara et al., 2004)(4). Interesting studies suggested that TCV-CP interacts with a protein belongs to the NAC family of transcription activators, which is designated TIP (TCV interacting protein) and prevents nuclear localization of TIP (Ren et al., 2005)(12). These observations suggested that TIP acts as a guardee protein, which imply that HRT is activated when CP alters the cellular distribution of TIP. However, we found that the mutation in TIP does not compromise in HRT-mediated resistance to TCV, but does compromise basal resistance to TCV and Cucumber mosaic virus (Jeong et al., 2008)(24). In addition, HRT protein is localized to the plasma membrane. This was confirmed using transgenic lines expressing HRT-GFP under the HRT promoter, where HRT-GFP was localized exclusively to the periphery of the cell, compared to GFP alone which was distributed uniformly throughout the cell.
Role of SA in HRT-mediated resistance signaling to TCV
Exogenous application of SA can confer resistance in HRT RRT, but not in hrt RRT plants. This is because, SA upregulates HRT expression, which can overcome the requirement for rrt (Chandra-Shekara et al., 2004)(4). SA-mediated upregulation of HRT also confers extreme resistance to TCV, wherein viral replication is restricted to the initial few cells, resulting in microscopic HR (Chandra-Shekara et al., 2004; Jeong et al., 2008)(4, 24). This indicates that HRT-mediated resistance is dependent on SA but not on JA and ethylene (Chandra-Shekara et al., 2004; Kachroo et al., 2000)(4, 5). SA is able to synthesized either from phenylalanine via the phenylalanine ammonia lyase (PAL) or from chorishmate via the shikimate pathway in plants (Wildermuth et al., 2001)(27). The small phenolic compound SA plays a critical role in the disease resistance signaling to different pathogens.
Arabidopsis mutants that are impaired in SA responsiveness, such as npr1 (nonexpressor of PR), or are defective in pathogen-induced SA accumulation, such as eds1 (enhanced disease susceptibility), eds5, sid2 (isochorishmatesynthase) and pad4 (phytoalexin deficient 4) confer enhanced susceptibility to a variety of plant pathogens (Nawrath et al., 2002; Wildermuth et al., 2001)(26, 27). TCV inoculation induces ~10-fold increase in SA levels in Di-17, but not in Col-0, which correlates with high expression of PR genes in Di-17 plants. Mutations that compromise SA biosynthesis (sid2 and eds5; Nawrath et al., 2002; Wildermuth et al., 2001)(26, 27) or SA signaling (eds1, pad4, sag101; Fey et al., 2005)(28) impair HRT-mediated resistance to TCV (Fig. 3). Unlike resistance, HR is not affected by sid2, eds5, eds1 or pad4 mutations, suggesting that HR and resistance mutually follow different pathways. EDS1, but not PAD4, acts redundantly with SA to regulate HRT-mediated signaling (Venugopal et al., 2009)(29). TIR-NBS-LRR types of R protein pathway mostly require the EDS1, while CC-NBS-LRR types of R protein pathway utilize NDR1. However, interestingly, HRT protein, which is a CC-NBS-LRR type R protein is dependent on EDS1 but independent of NDR1 (Chandra-Shekara et al., 2004)(4).
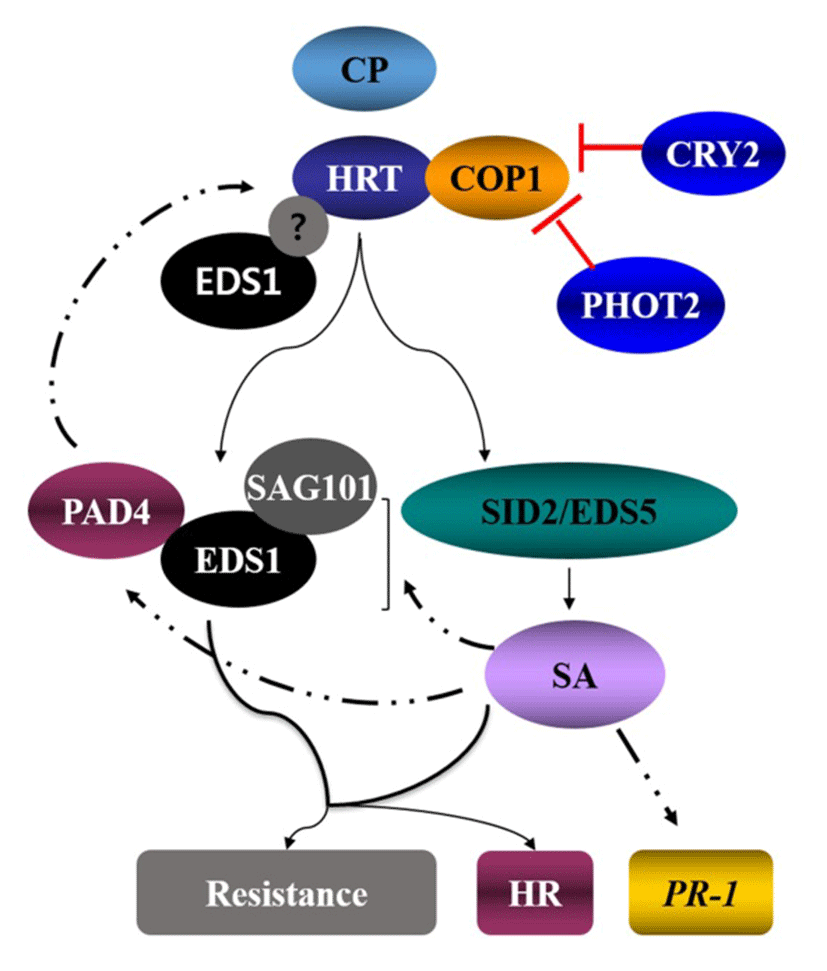
Some ecotypes of Arabidopsis contain two functional isoforms of EDS1 and these appear to be functionally redundant (Zhu et al., 2011)(30). EDS1 forms two distinct complexes with PAD4 and SAG101 since direct interaction between PAD4 and SAG101 has not been detected. EDS1 and PAD4 are localized in the nucleus and cytoplasm, whereas SAG101 is dominantly present in the nucleus. A recent study indicated that the EDS1, PAD4 and SAG101 also form a ternary complex and the localization of protein complexes depends on the relative compartmentalization of these three proteins (Zhu et al., 2011)(30). EDS1, but not PAD4 or SAG101, directly influences HRT-CP-mediated cell death response. Both isoforms of EDS1 interact with HRT, suggesting that EDS1 might participate in the CP-mediated activation of HRT (Zhu et al., 2011)(30).
Role of light in HRT-mediated resistance signaling to TCV
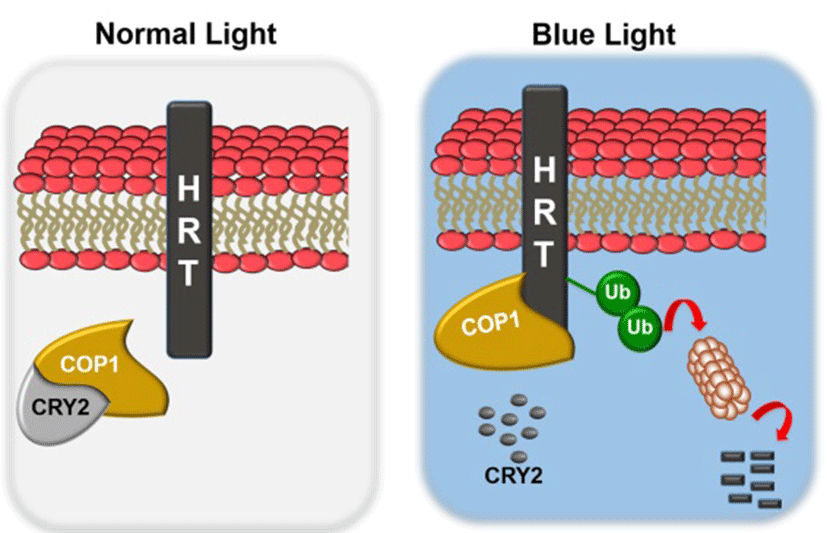
Light is also required for HR and resistance to TCV, since a dark period immediately following TCV inoculation abolishes both HR and resistance (Chandra-Shekara et al., 2006)(31). This suggests that light plays a critical role in HRT-mediated resistance. From the further study, mutations in the blue-light photoreceptors (CRY1, CRY2, PHOT1, and PHOT2), but not red and far-red photoreceptors (PHYA, PHYB, PHYC, PHYD, and PHYE) (Chen et al., 2004)(32), impaired resistance to TCV. In case of other pathosystem, phyA or phyB compromise the ability to induce localized cell death at the site of pathogen entry and also repressed in the SA-induced expression of the PR genes. Moreover, recent studies suggested that PHYA PHYB is compromised for systemic acquired resistance (SAR) (Griebel et al., 2008)(33). In addition to red-light photoreceptors, blue-light photoreceptors are known to participate in plant defense to bacterial pathogen. For example, CRY1 positively regulates RPS2-mediated resistance and SAR under continuous light (Wu et al., 2010)(34). Analysis of HRT protein levels in cry1, cry2, phot1 or phot2 plants showed significant reductions in HRT levels in cry2 and phot2 plants, but not in cry1 or phot1 plants. Thus, the CRY2 and PHOT2 proteins likely contribute to HRT-mediated resistance by affecting the stability of the HRT protein. Interestingly, exogenous SA was able to enhance TCV resistance in HRT cry1, HRT phot1 and HRT phot2 plants, but not in HRT cry2 plants. HRT stability also correlated with the presence of CRY2, which is degraded in a blue-light specific manner; exposure to blue-light resulted in degradation of both CRY2 as well as HRT. Pretreatment with the 26S proteasome-specific inhibitor, MG132, significantly inhibited the blue light-mediated degradation of the HRT-FLAG protein. Consistent with these results, HRT interacted with the CRY2- and PHOT2-interacting protein, COP1 (Jeong et al., 2010)(35), an E3 ubiquitin ligase involved in 26S proteasome- mediated degradation of proteins. This finding raised hypothesis that under normal light conditions, CRY2 and/or PHOT2 repress COP1-mediated degradation of HRT. On the other hand, in the dark, or under blue-light conditions, CRY2 degradation and possible conformation changes in PHOT2 relieve the repression of COP1 activity. Thereby this enables COP1 to interact with HRT and thus affects HRT degradation (Fig. 3 and 4).
RNA silencing components mediate resistance signaling against TCV
In the absence of the R protein, plants can induce a basal defense response against pathogens, although this mode of defense fails to contain the pathogen in contrast to R gene-mediated resistance. Basal defense to viruses often involves RNA silencing (reviewed in Ding, 2010; Voinnet, 2010)(36, 37). RNA silencing is induced upon the formation of a double stranded (ds) RNA, which is processed to small (s) 20-30 nucleotide (nt) dsRNA with staggered ends. One strand of the dsRNA is then incorporated into a large ribonucleoprotein complex called the RNA-induced silencing complex (RISC), which then cleaves the target viral RNA. The ribonuclease III enzymes called Dicers, mediate the processing of dsRNA into sRNA. Arabidopsis plants encode four Dicer-like proteins (DCL1 to DCL4) and of these DCL2, DCL3, and DCL4 process long dsRNA molecules of various cellular origins into sRNA that are 22, 24 or 21 nt in length, respectively. RISC complexes are formed of sRNA strand and a member of the Argonaute (AGO) protein family, which are also called slicer proteins since they cleave target single stranded RNA at the duplex formed with the guide-strand sRNA. There are ten AGO homologues in Arabidopsis, but the functions of most of these remain unclear. Although emerging results implicate small RNA in R protein-mediated resistance (Navarro et al., 2006; Bhattacharjee et al., 2009)(38, 39), the relationship between R protein-mediated signaling and RNA silencing pathway remains unclear. Viruses in turn have evolved to express suppressors that target host RNA silencing components and thereby ensure replication in the host. Interestingly, in many cases these suppressors of silencing also acts as avr factors and their direct/indirect interaction with the host R proteins leads to activation of defense responses.
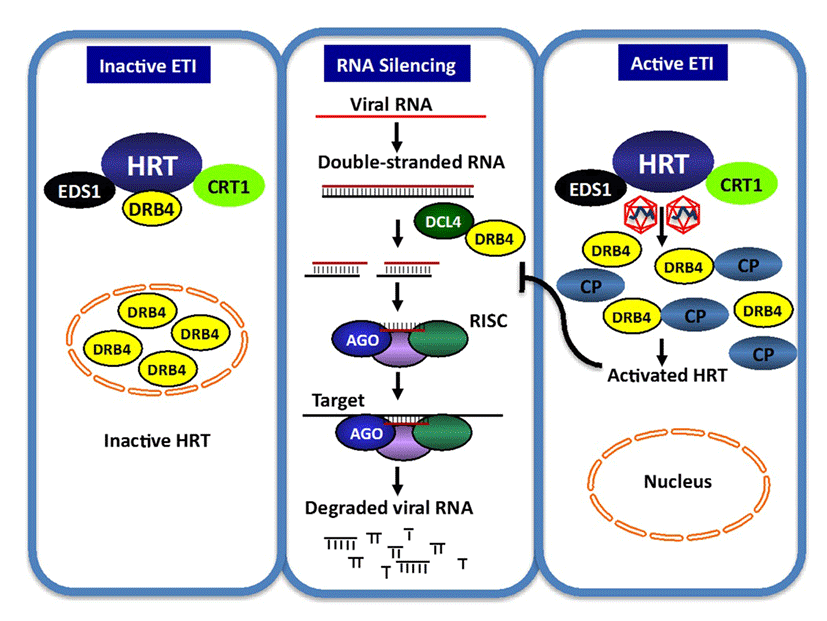
Components of the Arabidopsis RNA silencing machinery, including RDR6, DCL4, and DRB4, are required for HRT-mediated resistance and of these HRT, DRB4 and DCL4 are present in a single complex. HRT forms a complex, involving inter- and intra-molecular interactions with itself as well as with several other host proteins including DRB4 and DCL4 (Zhu et al., 2013)(40). The pathogen encoded avr effector, coat protein, specifically prevents HRT-DRB4 interaction and consequently HRT-DRB4-DCL4 complex formation. Notably, CP does not disrupt interactions between DRB4-DLC4 or HRT-HRT. TCV CP mutant R8A, which is able to induce HRT-mediated HR and PR-1 expression, can also prevent HRT-DRB4 interaction. However, unlike wild-type TCV, R8A virus remains virulent on Di-17 plants. This suggests that the dissociation of the HRT-DRB4 complex alone may not be sufficient for the activation of HRT. The fact that DRB4 directs CP, but not the R8A and R6A CP mutants, to the nucleus, correlates with the loss of avirulence in the R8A and R6A mutants on Di-17 plants. This result further suggests that the nuclear localization of CP might be important for the resistance response. The reciprocal ability of CP alone to redirect DRB4 to the nucleus and periphery further suggests that the resistance response might involve the redistribution of cellular proteins, and that the combined effects of these events might contribute to prevent the spread of the virus (Fig. 5).
Conclusions
To date, plant viruses are still a major threat to the crops and causing serious economic losses worldwide. Several decades ago, viral pathogens have been mostly controlled by conventional and chemical protection approaches. To further control of plant viral disease, tremendous progress has been made towards improving our understanding of plant viruses and the ways in which plants defend themselves against these pathogens. Our ever- increasing knowledge of the molecular mechanisms that restrict or allow viral infection has successfully been used to develop antiviral strategies. Despite a great deal of recent insights into the mechanisms by which R proteins recognize pathogens in general, and viruses in particular, there are still many gaps in our knowledge of how these proteins function and a great deal of room for further study. Understanding resistance associated signal transduction pathway to TCV and characterizing key genes involved in TCV resistance will be very useful in engineering virus resistant crops. Further exploitation of HRT-mediated resistance mechanisms, as well as understanding with unknown defense signaling, will provide new potential strategies to viral pathogens. In turn, these insights may lead to better strategies for control of diseases caused by viruses and other plant pathogens.






